Virtual Chemoproteomics Platform
Covalent fragment libraries in proteomic screenings are gaining popularity for lead finding. The molecules are typically larger than the preferred size of reversible fragments due to built-in spacers and warheads (except for MiniFrags). The increased complexity, the larger number of heavy atoms of the library compounds as well as the spacial and electrostatic requirements contribute to a much larger chemical space that need to be covered significantly lowering the expected hit-rate. We have developed a virtual chemoproteomics platform to sample a much larger chemical space and increase the success-rate for lead identification against hard targets.
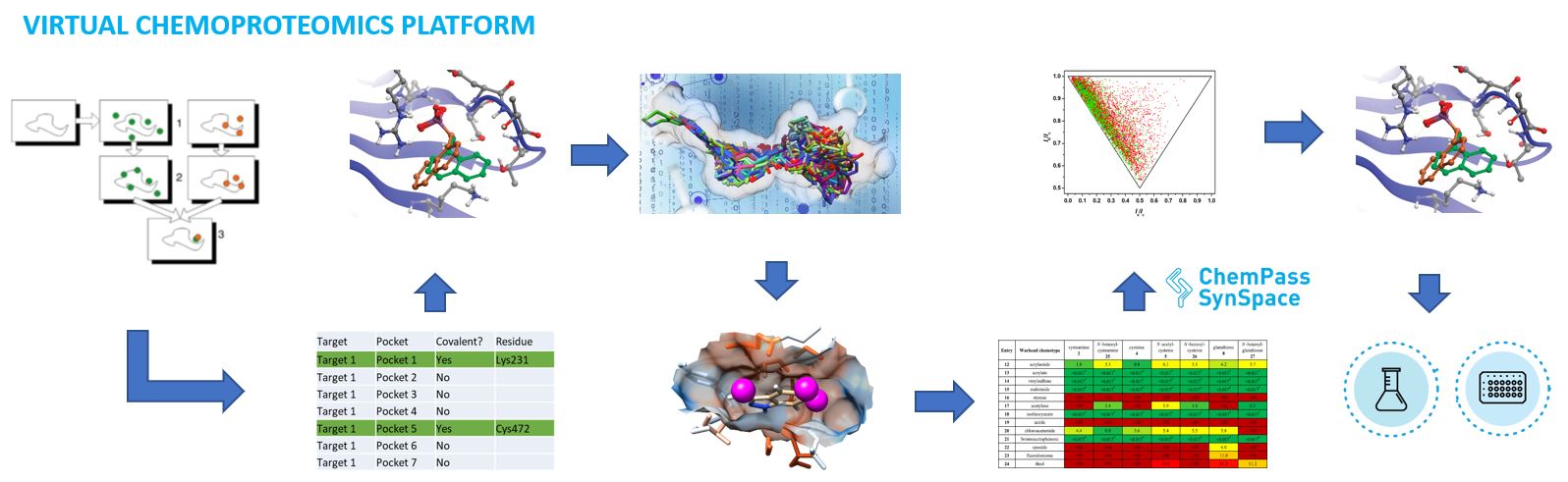
The platform first identifies all pockets of interest in the target protein and evaluates the the accessibility of the nucleophilic atoms of covalently modifiable amino acids within and in the vicinity of the pockets. Exhaustive virtual screening of all small reversible fragments ensure the coverage of a very large fragment chemical space to obtain noncovalen fragment binders. The proprietary decoy analysis and fragment growing tools generate a large focused covalent fragment set varying spacers and warheads suitable for each tragetable residue. Finally, virtual covalent screening yield a small focused fragment set for each pocket. Due its exhaustive and strategic search methodology, the platform covers a much larger complexity space than any wet lab library can increasing the likelihood of success against hard targets.
Virtual or confirmed covalent fragment hits can be subjected to a rapid optimization using the automated generative fragment growing and generative fragment optimization modules. The fragment growing tool automatically analyses the pocket around the ligands for growth vectors and substitution points before filling holes in a generative manner. The promising new enlarged fragment motifs are then subjected to generative fine-tuning to yield leads and advanced leads typically in just a 1 – 2 cycles. Due to the automated engines we can carry out optimization on a large number of fragments simultaneously and select only the best candidates for synthesis. All generative design steps are executed in synthetically feasible chemcial space that ensures short synthetic timelines, low cost and high laboratory success rates.
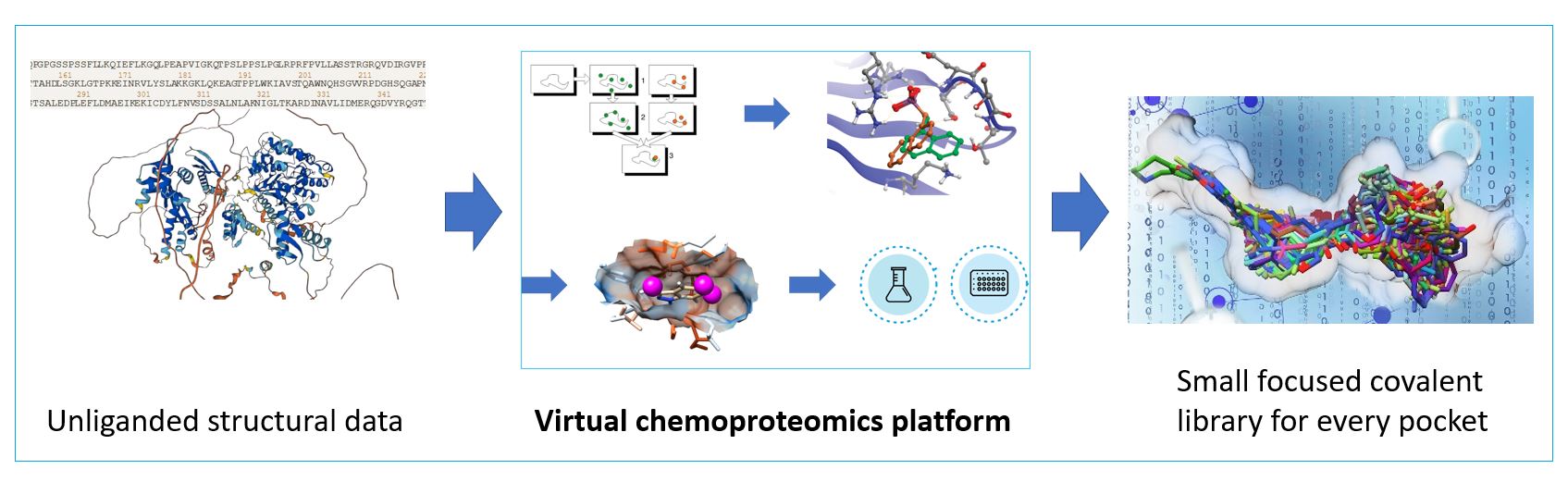
The platform first identifies all pockets of interest in the target protein and evaluates the the accessibility of the nucleophilic atoms of covalently modifiable amino acids within and in the vicinity of the pockets. Exhaustive virtual screening of all small reversible fragments ensure the coverage of a very large fragment chemical space to obtain noncovalen fragment binders. The proprietary decoy analysis and fragment growing tools generate a large focused covalent fragment set varying spacers and warheads suitable for each tragetable residue. Finally, virtual covalent screening yield a small focused fragment set for each pocket. Due its exhaustive and strategic search methodology, the platform covers a much larger complexity space than any wet lab library can increasing the likelihood of success against hard targets.
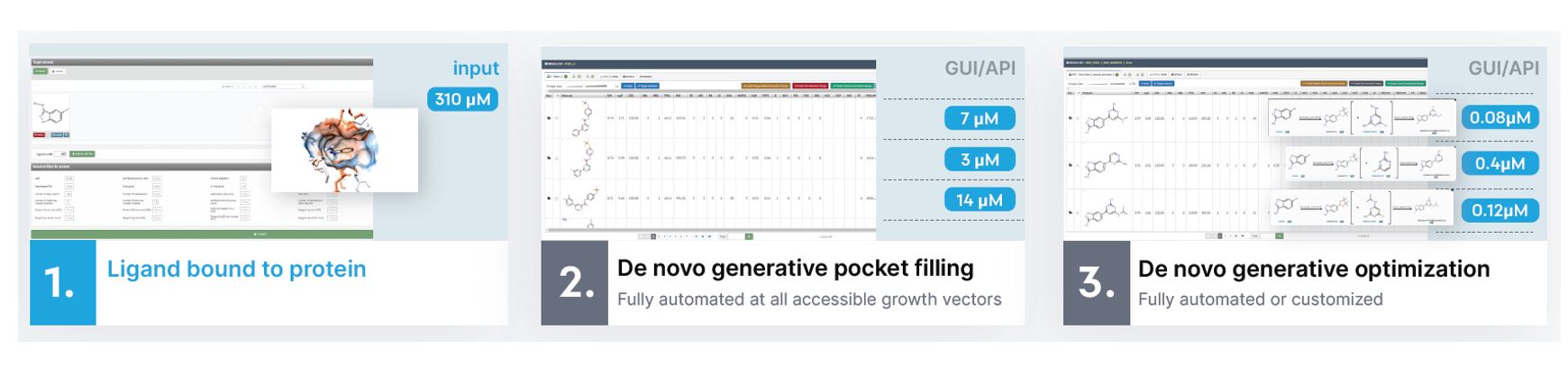
Virtual or confirmed covalent fragment hits can be subjected to a rapid optimization using the automated generative fragment growing and generative fragment optimization modules. The fragment growing tool automatically analyses the pocket around the ligands for growth vectors and substitution points before filling holes in a generative manner. The promising new enlarged fragment motifs are then subjected to generative fine-tuning to yield leads and advanced leads typically in just a 1 – 2 cycles. Due to the automated engines we can carry out optimization on a large number of fragments simultaneously and select only the best candidates for synthesis. All generative design steps are executed in synthetically feasible chemcial space that ensures short synthetic timelines, low cost and high laboratory success rates.
Universal Fragment Library Design Platform
The success of chemoproteomic screenings depends on the screening technique and quality of the screening libraries. Existing covalent and noncovalent fragment libraries are typically selected by diversity analysis of available fragments that possess desirable properties. Our fragment library design platform is an assembly of several key technologies required to revolutionize the design of fragment libraries. We deliver exclusive in silico libraries on a first-come-first-serve basis.
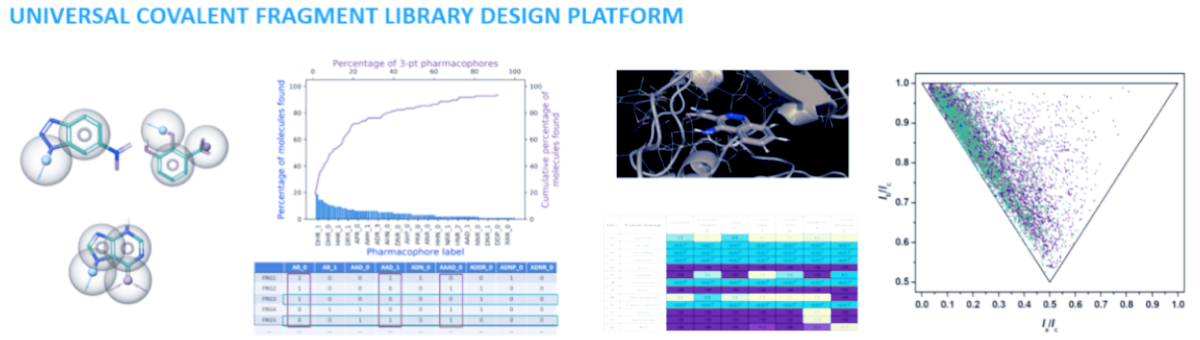
ChemPass in collaboration with Professor Gyorgy Keseru’s laboratory has developed all the necessary technologies that enable us to revolutionize fragment library design via our new platform. Instead of traditional diversity analysis, our universal fragment design platform evaluates the combinations of all experimentally observed pharmacophore multiplets and growth vectors as well as synthetic feasibility, warhead reactivity, and spacer diversity for covalent fragments sets. The result is a universally applicable covalent or noncovalent fragment library that delivers good success in both easy and challenging screening campaigns. Due to the built-in customization capabilities of the platform, the library design can target specific protein classes, and the library size and the synthetic cost can also be minimized. The concept has been validated by screening a test library against a validated and a hard target producing a hit rate and binding site coverage against the hard target much superior than results obtained with a standard fragment library.
Traditional fragment libraries designed by diversity analyses have been shown to include a large number of redundant pharmacophore and spacer motifs resulting in high hit rates of redundant rings for certain targets and few-to-no hits against pockets of difficult targets. Classical fingerprints do not work well with tiny fragment size molecules and fingerprints in general represent pharmacophore concepts poorly. Furthermore, potential growth vectors for warheads or substituents are not taken into account by traditional library design approaches despite evidence that most binding sites will only accommodate a few growth vectors. The overall result is a highly variable experimental hit rate and high risk of missed opportunities.
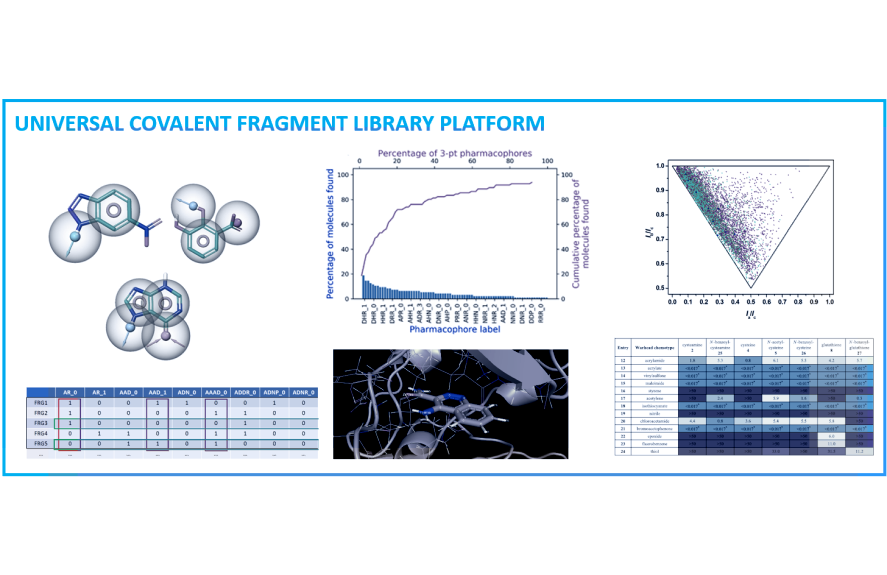
ChemPass in collaboration with Professor Gyorgy Keseru’s laboratory has developed all the necessary technologies that enable us to revolutionize fragment library design via our new platform. Instead of traditional diversity analysis, our universal fragment design platform evaluates the combinations of all experimentally observed pharmacophore multiplets and growth vectors as well as synthetic feasibility, warhead reactivity, and spacer diversity for covalent fragments sets. The result is a universally applicable covalent or noncovalent fragment library that delivers good success in both easy and challenging screening campaigns. Due to the built-in customization capabilities of the platform, the library design can target specific protein classes, and the library size and the synthetic cost can also be minimized. The concept has been validated by screening a test library against a validated and a hard target producing a hit rate and binding site coverage against the hard target much superior than results obtained with a standard fragment library.
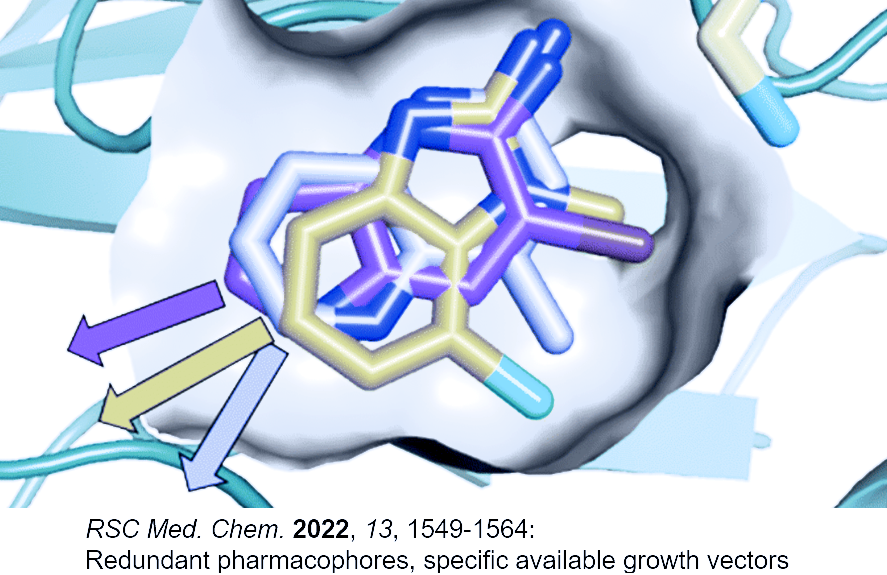
Traditional fragment libraries designed by diversity analyses have been shown to include a large number of redundant pharmacophore and spacer motifs resulting in high hit rates of redundant rings for certain targets and few-to-no hits against pockets of difficult targets. Classical fingerprints do not work well with tiny fragment size molecules and fingerprints in general represent pharmacophore concepts poorly. Furthermore, potential growth vectors for warheads or substituents are not taken into account by traditional library design approaches despite evidence that most binding sites will only accommodate a few growth vectors. The overall result is a highly variable experimental hit rate and high risk of missed opportunities.